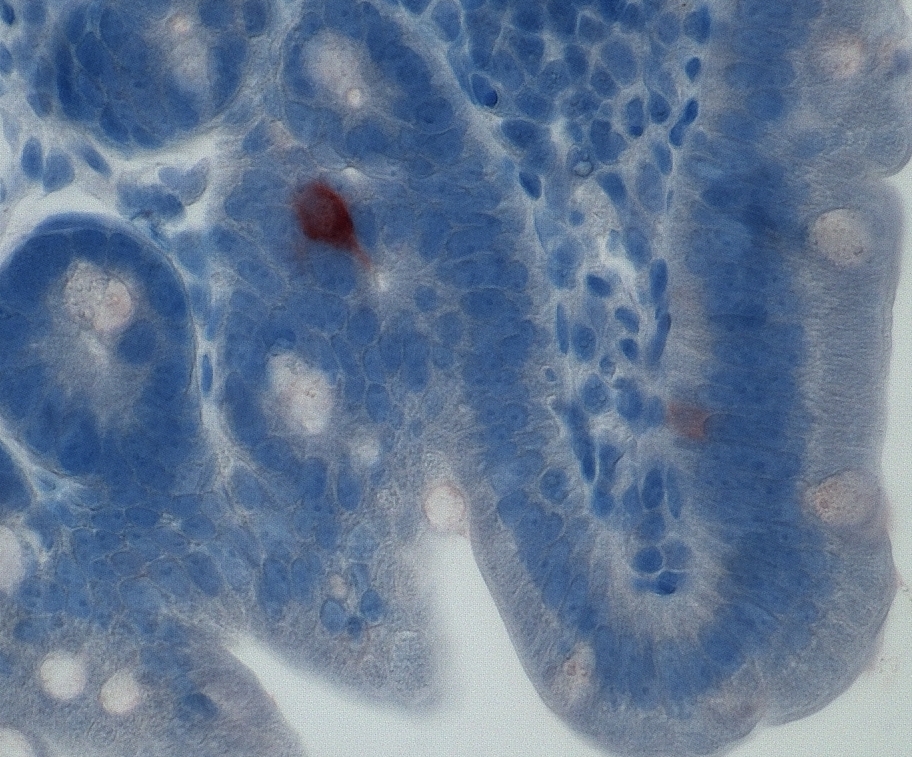
Reprogrammed mouse intestinal cells secreting insulin (red)
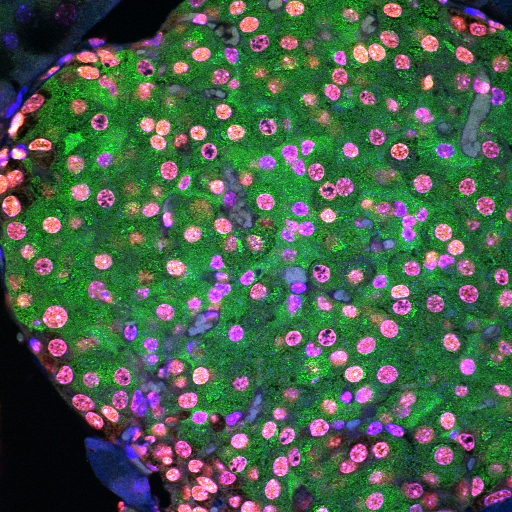
Rat pancreas stained for insulin (green) and PDX-1 (red)
Human carcinoma being attacked by enteric pathogen
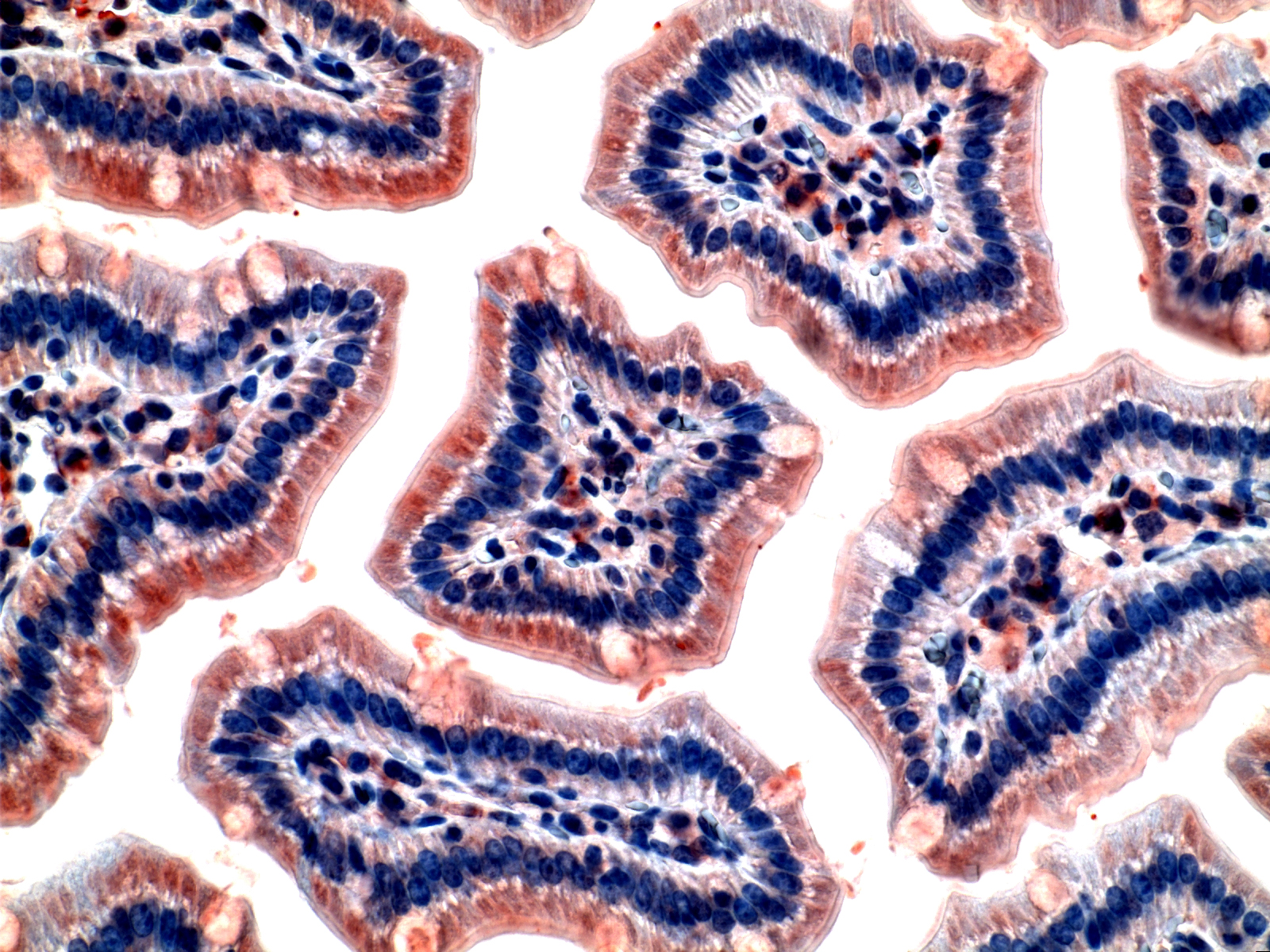
Mouse intestinal cells stained for cholera toxin
Uncut villous scaffolds prior to permeabolizing
Welcome to the Metabolic and Signal Engineering Labs of John C. March. We are located in the Department of Biological and Environmental Engineering at Cornell University. This web page is intended to tell you more about our research in metabolic and signal engineering. Signaling is how cells communicate with one another and with the world around them.